Clinical Bacteriology Testing at DSHS Laboratory
Texas Notifiable Conditions Surveillance
The clinical bacteriology group receives and tests many of the specimens submitted for notifiable conditions. The Texas Notifiable Conditions List is available at Notifiable Conditions | Texas DSHS.

2024 Texas Notifiable Conditions List
The process of reporting a notifiable condition to DSHS includes
- Reporting the condition to the local or regional health department (of the patient’s county of residence) within the reporting timeframe.
- Submitting an isolate or clinical specimen to the DSHS Laboratory. This is a required step for some, but not all notifiable conditions.
Texas Admin. Code 25 §97.2 identifies who must notify the local health authority of a person or animal suspected of having a notifiable condition.
Texas Admin. Code 25 §97.3 (4) identifies what specimens of these must be submitted to DSHS Laboratory for surveillance.
View an outline of the reporting and specimen submission process.
Note: Only some notifiable conditions require a specimen be sent to the DSHS Laboratory. The table below lists the notifiable condition specimens processed by the Clinical Bacteriology Group only. Refer to the Notifiable Conditions List for the complete list of required specimens. Pure cultures of the organisms should be submitted as they become available.
| Specimen | Submission Form Type | Select in Section 4.1 | Notes |
|---|---|---|---|
| Corynebacterium diphtheriae isolates | G-28 | Corynebacterium diphtheriae |
|
| Haemophilus influenzae isolates from normally sterile sites in children younger than 5 years old | G-28 | Haemophilus influenzae |
|
| Listeria monocytogenes isolates | G-28 | Listeria |
|
| Neisseria meningitidis isolates from normally sterile sites or purpuric lesions | G-28 | Neisseria meningitidis |
|
| Salmonella species isolates, including Salmonella positive specimens by CIDT | G-28 | Salmonella |
|
| Shiga toxin producing Escherichia coli (All E. coli O157:H7 isolates and any E. coli isolates or specimens in which Shiga toxin activity is demonstrated) | G-28 | Shiga toxin producing Escherichia coli |
|
| Staphylococcus aureus with a vancomycin MIC greater than 2 μg/mL (VISA and VRSA) | G-28 | Staphylococcus aureus |
|
| Streptococcus pneumoniae isolates from normally sterile sites in children younger than 5 years | G-28 | Streptococcus pneumoniae |
|
| Vibrio species isolates and Vibrio positive specimens (by CIDT) | G-28 | Vibrio sp. |
|
Diphtheria
Diphtheria is a potentially serious respiratory and skin disease caused by toxin producing Corynebacterium diphtheriae bacteria. The bacteria are usually spread from person-to-person via respiratory secretions. Occasionally it may spread through contact with skin lesions. Confirmed and suspected cases of diphtheria are immediately reportable in Texas.
via respiratory secretions. Occasionally it may spread through contact with skin lesions. Confirmed and suspected cases of diphtheria are immediately reportable in Texas.
Diphtheria Testing at DSHS Austin Laboratory
The Laboratory carries out isolation and identification of Corynebacterium diphtheriae.
Please refer to DSHS’ LTSM test menu for more details on C. diphtheriae testing at the Laboratory. General specimen submission guidance is available on DSHS’ specimen submission and shipping requirements pages.
For more information on diphtheria, visit the DSHS’ Infectious Disease Control’s webpages at Diphtheria | Texas DSHS and CDC at Diphtheria | CDC.
Haemophilus influenzae Disease
Haemophilus influenzae disease is the name for any disease caused by six identifiable types of H. influenzae bacteria (types a through f) or non-typeable H. influenzae bacteria. H. influenzae, type b (Hib) usually causes the most severe disease and is the only type that is vaccine preventable. Some of these illnesses, such as ear infections, are mild while others, such as bloodstream infections, are invasive and serious. Despite its name, H. influenzae does not cause influenza (the flu). All cases of invasive H. influenzae disease are reportable in Texas.
non-typeable H. influenzae bacteria. H. influenzae, type b (Hib) usually causes the most severe disease and is the only type that is vaccine preventable. Some of these illnesses, such as ear infections, are mild while others, such as bloodstream infections, are invasive and serious. Despite its name, H. influenzae does not cause influenza (the flu). All cases of invasive H. influenzae disease are reportable in Texas.
The Laboratory carries out identification of H. influenzae isolates. Specimens of invasive H. influenzae infections (taken from normally sterile sites such as blood or CSF) in children younger than 5 years must be submitted to DSHS for testing. Specimens from non-sterile sites (such as swabs) are not acceptable as notifiable condition specimen submissions. Specimens from older patients are not required for reporting purposes.
Haemophilus influenzae Testing at the DSHS Austin Laboratory
Please refer to the DSHS’ LTSM test menu for more details on H. influenzae testing at the Laboratory. General specimen submission guidance is available on the DSHS’ specimen submission and shipping requirements pages.
For more information on Haemophilus influenzae disease, visit the DSHS’ Infectious Disease Control’s webpages at Haemophilus Influenzae (Including Hib) | Texas DSHS and CDC at Haemophilus influenzae Disease (including Hib) | CDC
Listeriosis
Listeriosis is a serious infection caused by eating food contaminated with Listeria monocytogenes bacteria. L. monocytogenes is most likely to sicken pregnant women and their newborns, adults aged 65 or older, and people with weakened immune systems. Listeriosis is a reportable condition in Texas.
by eating food contaminated with Listeria monocytogenes bacteria. L. monocytogenes is most likely to sicken pregnant women and their newborns, adults aged 65 or older, and people with weakened immune systems. Listeriosis is a reportable condition in Texas.
Listeria monocytogenes Testing at DSHS Austin Laboratory
The DSHS Laboratory carries out identification of L. monocytogenes isolates. The Laboratory also identifies Listeria species, including monocytogenes, from submitted food and environmental samples.
Please refer to the DSHS’ LTSM test menu for more details on L. monocytogenes testing at the Laboratory. General specimen submission guidance is available on the DSHS’ specimen submission and shipping requirements pages.
For more information on listeriosis, visit the CDC at Symptoms | Listeria | CDC
Meningococcal Invasive Disease
Meningococcal disease is a serious and potentially life-threatening infection caused by Neisseria meningitidis bacteria. N. meningitidis is spread from person-to-person either by:
potentially life-threatening infection caused by Neisseria meningitidis bacteria. N. meningitidis is spread from person-to-person either by:
- Direct contact with respiratory and throat secretions (e.g., kissing)
- Indirect contact (e.g., sharing of eating utensils, toothbrushes), or
- Aerosol droplets (e.g., coughing and sneezing).
Common signs and symptoms of meningococcal disease include sudden onset of high fever, neck stiffness, confusion, nausea, vomiting, lethargy, and rash. Without prompt and appropriate treatment, the infection can progress rapidly and result in death. All cases of meningococcal invasive disease are immediately reportable in Texas.
Neisseria meningitidis Testing at the DSHS Austin Laboratory
The Clinical Bacteriology Laboratory carries out identification of N. meningitidis isolates.
The Laboratory also serotypes N. meningitidis from sterile sites or purpuric lesions. Please refer to the DSHS’ LTSM test menu for more details on N. meningitidis testing at the Laboratory. General specimen submission guidance is available on the DSHS’ specimen submission and shipping requirements pages.
For more information on Meningococcal Invasive Disease, visit the DSHS’ Infectious Disease Control’s webpages at Meningococcal Invasive Disease | Texas DSHS and the CDC at Meningococcal Disease | CDC.
Salmonellosis
Salmonellosis is an intestinal illness caused by infection by Salmonella bacteria. It is spread by eating contaminated foods, drinking contaminated water, or ingesting feces of an infected person or animal. Salmonellosis is commonly acquired by:
- eating raw or undercooked poultry, eggs, or raw milk products or drinking raw milk or untreated water,
- caring for a person with diarrhea,
- working in a day-care facility or with diapered children,
- handling pets (including reptiles and poultry) and/or wild animals, or livestock or cleaning their living areas.
Salmonella spp. Testing at DSHS Austin Laboratory
The Clinical Bacteriology Laboratory screens stool specimens for Salmonella. Clinical specimens that are positive for Salmonella by culture independent diagnostic test (CIDT) methods are accepted for confirmation testing. ![]()
The DSHS Laboratory also tests food and environmental specimens for Salmonella.
Please refer to the DSHS’ LTSM test menu for more details on Salmonella testing at the Laboratory. General specimen submission guidance is available on the DSHS’ specimen submission and shipping requirements pages.
For more information on Salmonellosis, visit the DSHS’ Infectious Disease Control’s webpages at Salmonellosis (Salmonella Infection) | Texas DSHS and CDC at Salmonella Homepage | CDC.
Escherichia coli Infection
Escherichia coli is a large and diverse species of bacteria. They are found in the environment, foods, and intestines of people and animals. Most strains of E. coli are harmless, but some can cause illness, including diarrhea, urinary tract infections, and respiratory diseases.
Escherichia coli Testing at the DSHS Austin Laboratory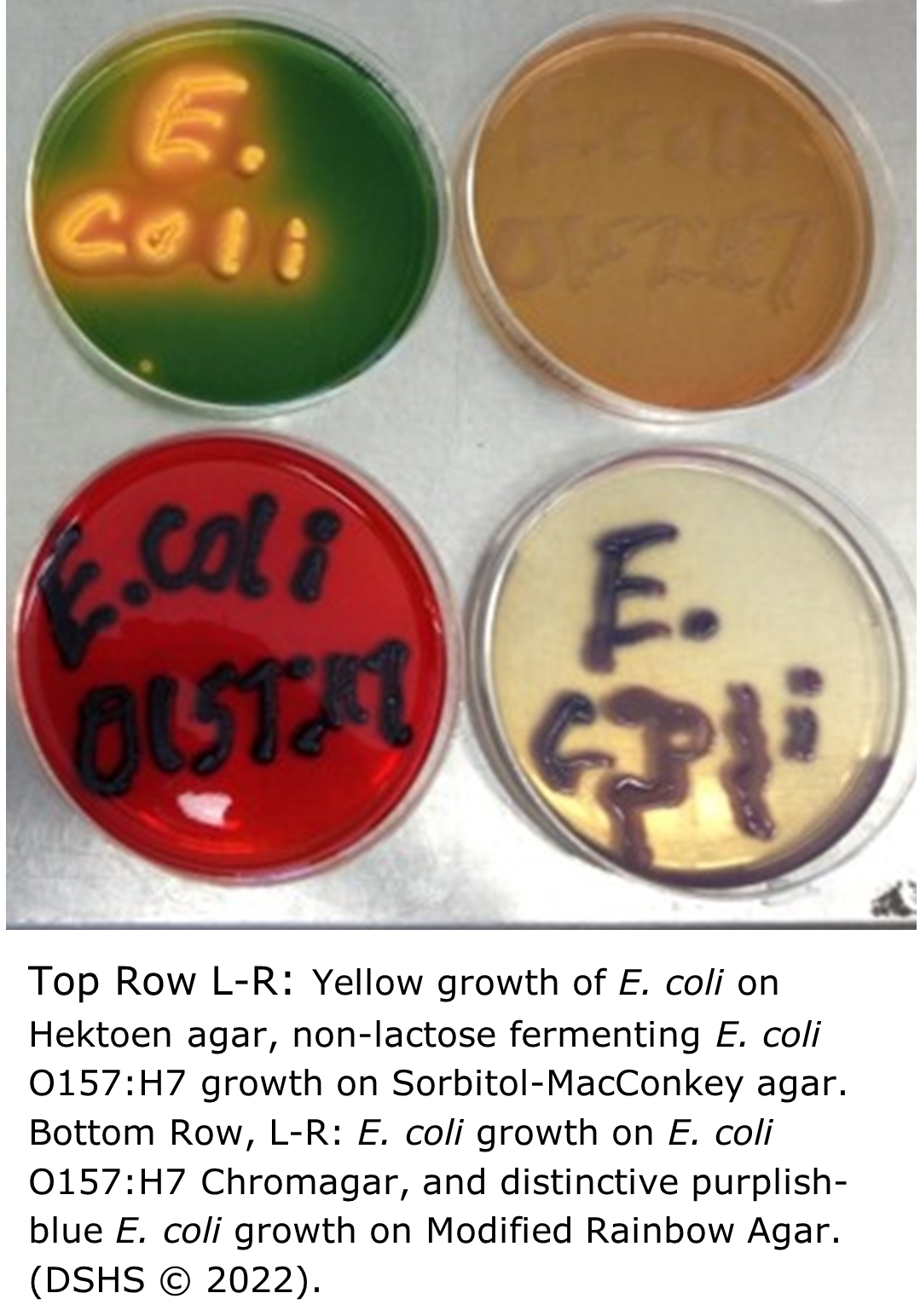
The Clinical Bacteriology Laboratory tests clinical stool specimens and pure isolate E. coli for Shiga-like toxins and Enterohaemorrhagic E. coli (EHEC).
The DSHS Laboratory also tests food and environmental specimens for Shiga-toxin producing E. coli, and tests drinking water for the presence of E. coli and other coliforms.
Please refer to the DSHS’ LTSM test menu for more details on E. coli and coliform testing at the Laboratory. General specimen submission guidance is available on the DSHS’ specimen submission and shipping requirements pages.
For more information on E. coli infection, visit the DSHS’ Infectious Disease Control’s webpages at E. coli Infection (Escherichia coli Infection) | Texas DSHS and CDC’s webpages at E. coli (Escherichia coli) | E. coli | CDC.
Note: Shiga toxin producing E. coli infection (all E. coli O157:H7 isolates and any E. coli isolates or specimens in which Shiga toxin activity has been demonstrated (STX1, STX2) are required to be submitted to the DSHS Laboratory per Texas Administrative Code.
Staphylococcus aureus “Staph” Infections
Staphylococcus aureus bacteria are commonly found on peoples’ skin and noses. Most of the time these bacteria are harmless, but some S. aureus can cause blood infections, pneumonia, bone and heart valve infections, and foodborne illnesses. People with chronic conditions or weakened immune systems, and those living in healthcare facilities, are at a higher risk of developing staph infections.
Staph infections are caused by several types of S. aureus. Many of these pathogenic S. aureus exhibit differing levels of resistance to antimicrobial compounds. Staphylococci of significant public health concern include:
- Methicillin-Resistant Staphylococcus aureus (MRSA)
- Methicillin-Susceptible Staphylococcus aureus (MSSA)
- Vancomycin-Intermediate Staphylococcus aureus (VISA)
- Vancomycin-Resistant Staphylococcus aureus (VRSA)
Staphylococcus aureus Testing at the DSHS Austin Laboratory
The Clinical Bacteriology Laboratory  identifies Staphylococcus and related species from fecal specimens and reference cultures. The antimicrobial susceptibility of Staphylococcus aureus specimens is also tested and determined, including susceptibility of isolates with a vancomycin MIC >2 μg/mL.
identifies Staphylococcus and related species from fecal specimens and reference cultures. The antimicrobial susceptibility of Staphylococcus aureus specimens is also tested and determined, including susceptibility of isolates with a vancomycin MIC >2 μg/mL.
The Laboratory also tests food and milk samples for the presence of staphylococcal enterotoxin.
Please refer to the DSHS’ LTSM test menu for more details on Staphylococcus identification and susceptibility testing at the Laboratory. General specimen submission guidance is available on the DSHS’ specimen submission and shipping requirements pages.
For more information on vancomycin resistant S. aureus, visit the DSHS’ Infectious Disease Control’s webpages at Vancomycin Intermediate Staphylococcus aureus (VISA) and Vancomycin Resistant Staphylococcus aureus (VRSA) | Texas DSHS. For information on S. aureus in healthcare settings, visit Staphylococcus aureus in Healthcare Settings | HAI | CDC.
Streptococcus pneumoniae Infection (Pneumococcal Disease)
 respiratory tract, nose, and sinuses of asymptomatic carriers. The bacteria can cause disease in certain conditions, usually if the host becomes immunocompromised. S. pneumoniae is the most common cause of community acquired pneumonia and meningitis in children and the elderly. The organism also causes many other types of infections other than pneumonia. Invasive pneumococcal diseases include bronchitis, sinus infections, blood infections, and ear infections. Invasive strep diseases are reportable in Texas.
respiratory tract, nose, and sinuses of asymptomatic carriers. The bacteria can cause disease in certain conditions, usually if the host becomes immunocompromised. S. pneumoniae is the most common cause of community acquired pneumonia and meningitis in children and the elderly. The organism also causes many other types of infections other than pneumonia. Invasive pneumococcal diseases include bronchitis, sinus infections, blood infections, and ear infections. Invasive strep diseases are reportable in Texas.
Streptococcus pneumoniae Testing at the DSHS Austin Laboratory
The Clinical Bacteriology Laboratory identifies Streptococcus pneumoniae from reference cultures and identifies beta hemolytic Streptococcus spp. (Group B Strep) from pure isolates. Group A Streptococcus is also identified by the same test.
Please refer to the DSHS’ LTSM test menu for more details on Strep identification and susceptibility testing at the Laboratory. General specimen submission guidance is available on the DSHS’ specimen submission and shipping requirements pages.
For more information on Streptococcus pneumoniae and pneumococcal diseases, visit the DSHS’ Infectious Disease Control’s webpages at Streptococcus pneumoniae Infection (Pneumococcal Disease) | Texas DSHS. For additional pneumococcal information, visit Pneumococcal Disease | CDC.
Clinical Bacteriology Testing Services
The table below lists many of the organisms processed by the Clinical Bacteriology Group. The estimated turnaround time (TAT) is based on the understanding that clinical specimens are high priority, but during an outbreak situation the identification and confirmation of referred pure cultures will be delayed until the outbreak activities are completed.|
Organism |
Testing Offered |
What and How to Submit |
Estimated Turnaround Time (TAT) for Results |
Additional Information |
|---|---|---|---|---|
|
Aerobic Bacteria |
Isolation and Identification |
Blood, blood culture bottles, CSF, tissues, lymph nodes, stool/feces, sputum (for Legionella only) |
7–21 days |
|
|
Identification |
Pure isolate on agar slant (room temp.) |
2–21days |
Refer to specific organisms on the submission form or include any testing already performed. |
|
|
Anaerobic Bacteria |
Isolation and Identification |
Submit anaerobically |
4–21 days |
|
|
Identification |
Pure anaerobic isolate; Submit anaerobically |
2–14 days |
|
|
|
Bacillus spp. |
Identification |
Pure isolate on agar slant (room temp. or refrigerated) |
3–21 days |
|
|
Campylobacter spp. |
Identification |
Pure isolate on agar slant. (refrigerated) |
3–21 days |
|
|
Isolation from clinical specimen (see culture, stool) |
Stool specimen in Cary-Blair or GN broth transport medium, on cold packs Blood in blood culture bottle |
3–21 days |
|
|
|
Corynebacterium spp. |
Identification (See Gram positive rod) |
Pure isolate on agar slant |
2–21 days |
Refer to shipping guidance in test page for shipping temperatures. |
|
Corynebacterium diphtheriae |
Identification and toxin testing |
Pure isolate on agar slant. (refrigerated) |
3–5 days |
|
|
Isolation and identification |
Swab in Stuart’s transport, Aime's, or Loeffler’s transport medium for Diphtheria screening. (refrigerated) |
3–5 days Positive results called to submitter |
If C. diphtheria is isolated, it will be submitted to CDC for toxin testing. |
|
|
Enteric bacteria |
Identification |
Pure isolate on agar slant (room temp. or refrigerated) |
2–21 days |
Enteric Bacterial Isolate – Definitive Identification | Texas DSHS |
|
Escherichia coli O157:H7 |
Identification and Serotyping; Shiga toxin testing |
Isolates: Pure isolate on growth medium (room temp. or refrigerated) Clinical: Raw stool specimen in Cary-Blair or GN broth transport medium, on ice packs |
2–21 days Serologically O157:H7
|
All E. coli O157:H7 isolates and EHEC isolates, or specimens (STX1 and STX2) are required to be submitted to the DSHS Laboratory per TAC §97.3. |
|
Shiga toxin producing E. coli (EHEC) |
Identification, Isolation, and Shiga toxin testing |
Pure isolate on growth medium (refrigerated) |
3–21 days |
All E. coli O157:H7 isolates and EHEC isolates, or specimens (STX1 and STX2) are required to be submitted to the DSHS Laboratory per TAC §97.3. |
|
Stool specimen (500 µL minimum) in in Cary-Blair or GN broth transport medium; < 7 days old refrigerated on ice packs; > 7 days old frozen at -70°C on dry ice |
||||
|
Gram Negative Rods |
Identification |
Pure isolate on agar slant |
3–21 days |
|
|
Gram Positive Rods |
Identification |
Pure isolate on agar slant |
3–21 days |
|
|
Haemophilus ducreyi |
Identification |
Pure isolate on chocolate agar slant |
5–15 days |
|
|
Isolation and identification |
Lesion scrapings, urogenital swab from infected area, transported on wet ice |
Negative 21 days Positive 7–21 days |
Haemophilus ducreyi Isolation and Identification | Texas DSHS |
|
|
Haemophilus influenzae |
Identification and serotyping |
Pure culture on agar slant (room temp.) |
2–14 days |
Only isolates from invasive cases (normally sterile sites) from children younger than 5 years old are required to be submitted to the DSHS Laboratory per TAC §97.3. |
|
Legionella species |
Identification |
Pure isolate on BCYE slant (refrigerated) |
4–21 days |
|
|
Isolation and identification |
Pleural fluid, tissue, sputum, bronchial washings, in sterile, leakproof container. < 48 hrs. from collection refrigerated on ice packs; > 48 hrs. freeze at -70°C on dry ice |
4–21 days |
||
|
Listeria species |
Identification |
Pure isolate on non-glucose containing agar slants |
5–10 days |
|
|
Isolation and identification |
Blood, CSF, amniotic fluid, placental tissue, fetal tissue; shipped on wet ice within 48 hours of collection |
Negative 21 days Positive 5–14 days called to submitter |
Refer to shipping guidance in test page for shipping temperatures. |
|
|
Listeria monocytogenes |
Identification |
Pure, viable isolate on suitable solid media slant |
5–10 days |
|
|
Neisseria gonorrhoeae |
Identification, antimicrobial susceptibility to azithromycin, cefixime, ceftriaxone, and ciprofloxacin |
Pure, viable isolate on chocolate II or GC base +1 % growth supplements agar slants Alternate: bacteria should be resuspended (≥4 McFarland) in trypticase soy broth (TSB) with 15%–20% glycerol and freeze immediately at -70°C. |
2–10 days |
|
|
Isolation and identification, antimicrobial susceptibility to azithromycin, cefixime, ceftriaxone, and ciprofloxacin |
Swab (Copan ESwab) from urethra, throat, oropharynx, rectum, vagina, cervix, conjunctiva |
4–14 days |
|
|
|
Neisseria meningitidis |
Serotyping |
Pure isolate on agar slant from sterile sites or purpuric lesion (room temp.) |
3–10 days |
|
|
Neisseria spp. |
Identification |
Pure isolate on agar slant that supports growth (room temp.) |
2–21 days |
|
|
Pneumococcus (Streptococcus pneumoniae) |
Typing |
Pure isolate on agar slant. (room temp. or refrigerated) |
Specimens are submitted to the Minnesota ARLN Laboratory. Average TAT is 7–10 days but is dependent upon volume of testing conducted. |
|
|
Salmonella spp. |
Identification |
Pure isolate on agar slant. (room temp. or refrigerated) |
3–21 days |
|
|
Isolation and Identification |
Clinical specimen: Cary Blair, GN Broth, etc. (For specimens in which a CIDT was positive, and isolation is not performed, please send the original material that yielded the positive result.) (refrigerated or frozen at -70 °C) |
|||
|
Shigella spp. |
Identification |
Pure isolate on agar slant (refrigerated) |
2–21 days |
|
|
Isolation and Identification |
Clinical specimen: Cary Blair, GN Broth, etc. (For specimens in which a CIDT was positive, and isolation is not performed, please send the original material that yielded the positive result.) (refrigerated or frozen at -70°C) |
3–21 days |
|
|
|
Staphylococcus aureus |
Identification, Antimicrobial susceptibility testing for Vancomycin resistance (for epidemiological purposes only) |
Pure isolate on agar slant. |
3–7 days |
|
|
Isolation and Identification |
Stool specimens on ice packs (Outbreak situations only) |
2–4 days |
||
|
Staphylococcus species |
Identification |
Pure isolate on agar slant (room temp. or refrigerated) |
2–21 days |
|
|
Streptococcus species |
Identification |
Pure isolate on agar slant (room temp. or refrigerated) |
2–21 days |
|
|
Streptococcus group B |
Identification |
Pure isolate on agar slant (room temp. or refrigerated) |
2–21 days |
|
|
Vibrio cholerae |
Identification and serotyping |
Pure isolate on agar slant (room temp.) |
2–7 days Positive called to submitter within 48 hours |
|
|
Isolation, identification, and serotyping |
Stool specimen in Cary-Blair (room temp.) |
2–10 days Positive called to submitter |
||
|
Vibrio species |
Identification and serotyping |
Pure culture on agar slant (room temp.) |
4–7 days |
|
|
Isolation, identification, and serotyping |
Stool specimen in Cary-Blair on wet ice (room temp.) |
2–7 days Positive called to submitter |
||
|
Yersinia enterocolitica and other species |
Identification |
Pure cultures on agar slants |
4–7 days |
|
|
Isolation, identification |
Stool in Cary-Blair, Stuart, or Aime's transport, on wet ice |
4–7 days |